Abstract:
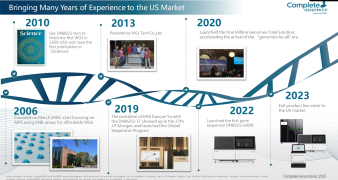
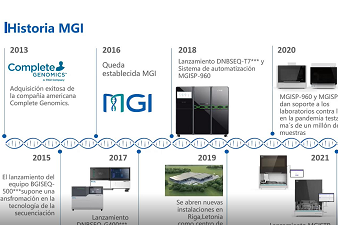
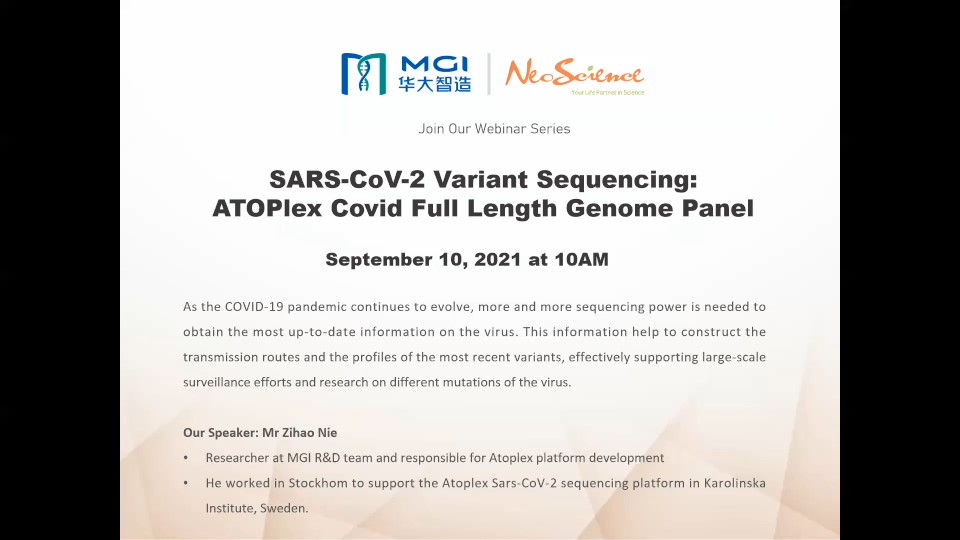
The ongoing COVID-19 pandemic caused by SARS-CoV-2 continues to emerge as a global health problem of unprecedented impacts. Here, we described MGI’s ATOPlex library workflow to obtain the SARS-CoV-2 genome libraries from randomly selected Nigerian patients that tested positive for COVID-19, including evaluating the performance of the DNBSEQ-G50 sequencer in SARS-CoV-2 lineage characterization. Analyses of the complete genome datasets of the SARS-CoV-2 isolates enabled us to obtain 293,258 – 20,000,000 raw reads with 291,502 - 19,879,520 (99.17 - 99.91%) mapping to the reference SARS-CoV-2 genome, and analysis revealed the occurrence of 9 distinct lineages among the SARS-CoV-2 isolates. To date, NIMR has sequenced hundreds more SARS-CoV-2 genomes from COVID-19 positive samples using the DNBSEQ-G50. These data will contribute to informing a robust and dynamic public health response for containment of the ongoing COVID-19 pandemic in Nigeria, Africa, and other continents of the world.
Speakers:
Prof. Iwalokun, Bamidele Abiodun - Director Central Research Laboratory (CRL), at Nigerian Institute of Medical Research (NIMR), Lagos – Nigeria
Dr. Ayorinde B. James - Molecular Biologist and Research Fellow, at Nigerian Institute of Medical Research (NIMR), Lagos – Nigeria
*Unless otherwise informed, StandardMPS and CoolMPS sequencing reagents, and sequencers for use with such reagents are not available in Germany, USA, Spain, UK, Hong Kong, Sweden, Belgium, Italy, Finland, Czech Republic, Switzerland, Portugal, Austria and Romania.
If you have any questions, you can send an email to:


 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL











 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club