CycloneSEQ™ Technology
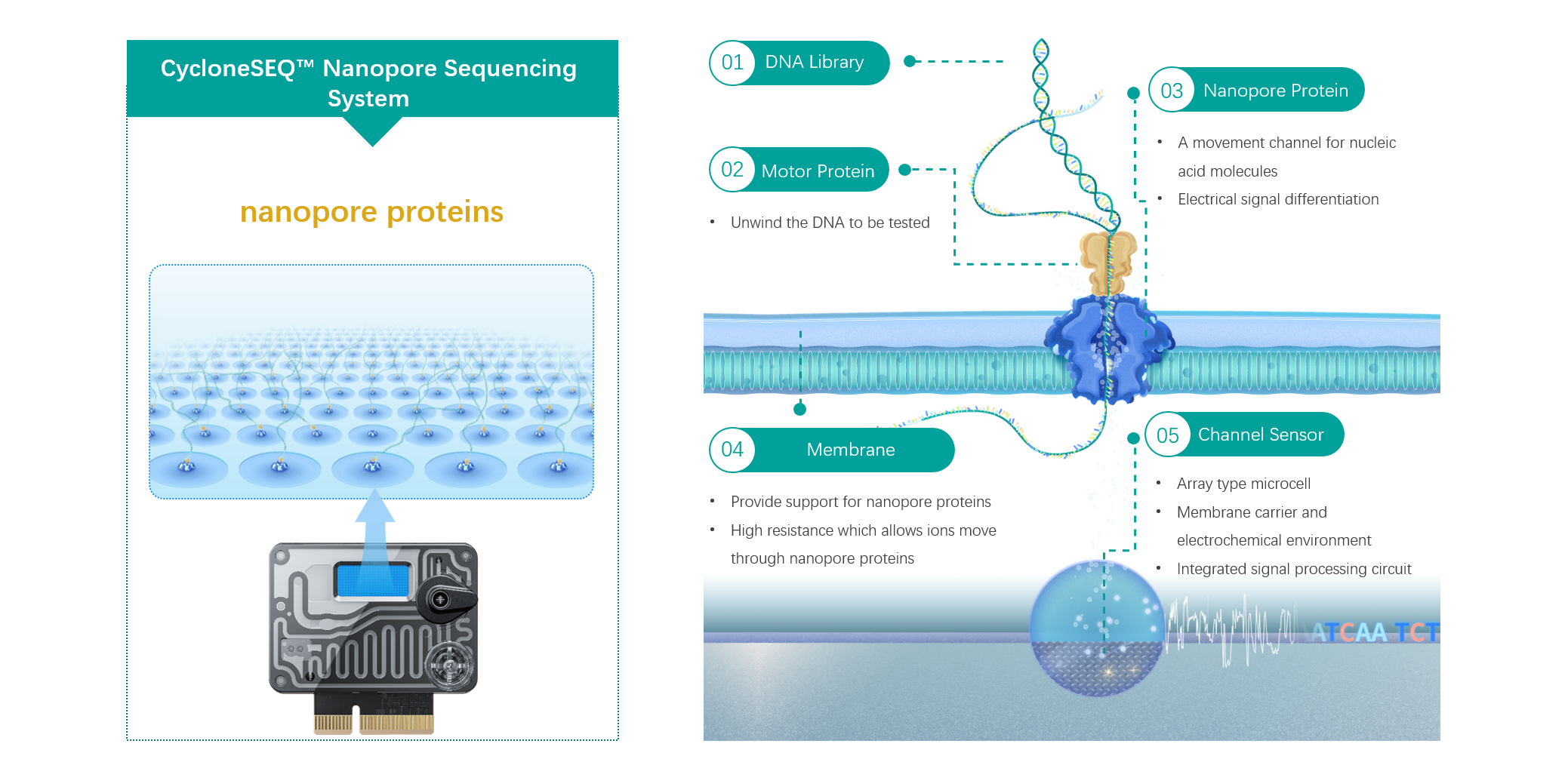
CycloneSEQ™ is a nanopore sequencing technology platform officially launched by the BGI Group in 2024. During the sequencing process of CycloneSEQ, the DNA library molecules linked to the motor proteins approach the entrance of the nanopore protein under the influence of an electric field. When the motor proteins reach the nanopore entrance, they are activated, effectively and rapidly unwinding the DNA, allowing the library to pass through the nanopore protein as single-stranded molecules. As different DNA bases and their arrangements obstruct the current to varying degrees, fluctuations in the current occur as the DNA passes through the nanopore. The channel sensors on the sequencing flow cell capture these current signals, which are then analyzed by the control software to obtain real-time information about the base sequence of the sample being tested.
CycloneSEQ™ and DNBSEQ™ sequencing technology can perfectly complement each other in terms of sequencing length and accuracy, enabling more comprehensive and precise analyses in various applications, such as: Genome Assembly, Detection of Genomic Structural Variations, Transcriptome Research.
Sequencing Principle of CycloneSEQ™
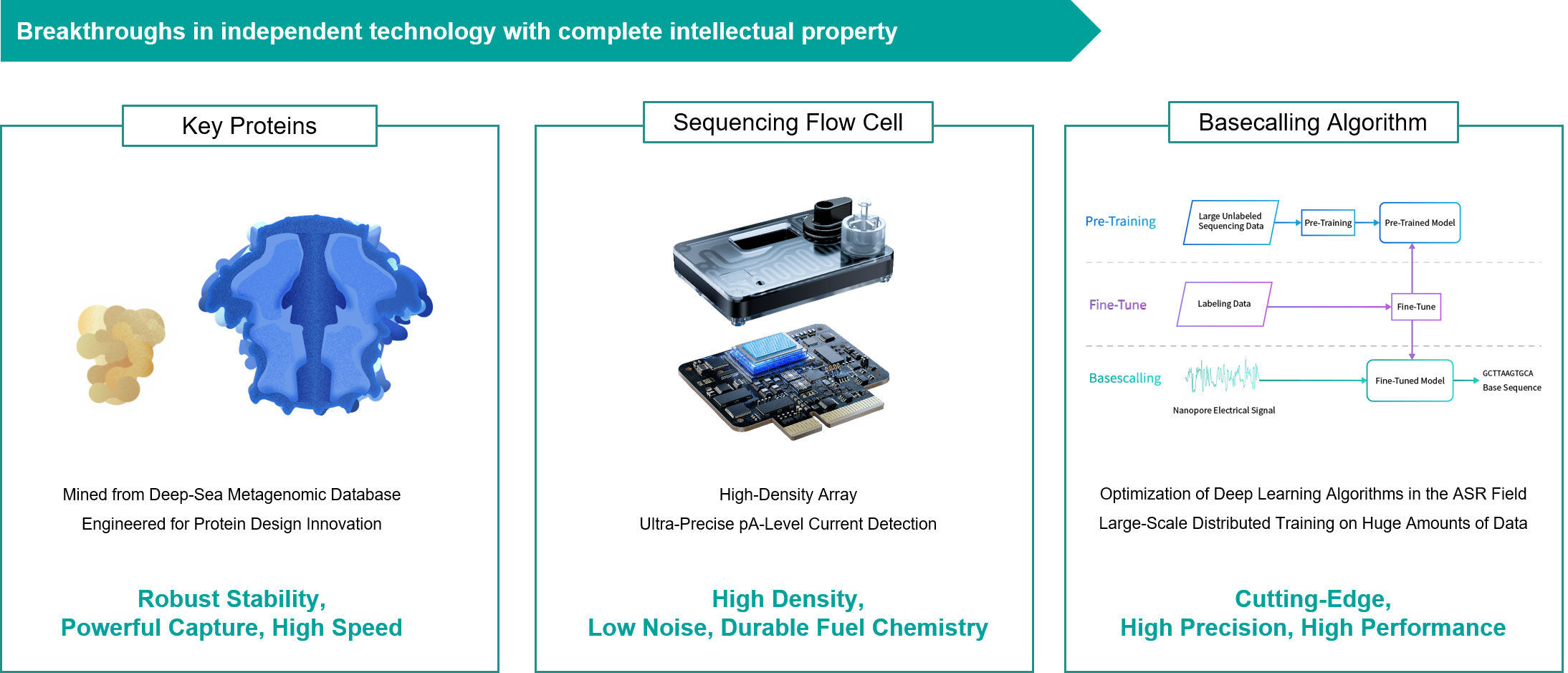
● Core Proteins: In the analysis of over 500 million deep-sea metagenomic databases collected from extreme environments such as the Mariana Trench, it was discovered that the deep sea has a high abundance of proteins related to genetic manipulation, with greater structural diversity. Among them, there is a highly robust nucleic acid manipulation protein, whose core functional region structure is significantly different from most known motor proteins, with stronger unspinning ability, and it is a source of hydrothermal region and has high thermal stability. Another is a densely structured pore protein formed by the self-assembly of homopolymers, characterized by strong structural rigidity, low sequencing noise, and high quality, with a small pore diameter. Through protein engineering, the motor protein has been designed to have fast sequencing speeds, strong stability, and the ability to sustain sequencing over long periods, while the pore protein features strong capture, high stability, sustained sequencing, and high signal-to-noise ratios, forming the core protein foundation of CycloneSEQ™.
● Core Flow cell: The nanopore protein sensors are arranged in a high-density array flow cell. The sensor microcavity diameter and spacing have been finely optimized for optimal membrane pore fitting, achieving ultra-low sequencing noise. The expanded microcavity volume allows for ultra-long sequencing durations. The integration of Bio Micro Electro Mechanical System (BioMEMS) with Application-Specific Integrated Circuits (ASIC) enables the precise detection of picoampere-level currents, providing real-time and stable reading of nucleic acid sequence information.
● Core Algorithms: Incorporating state-of-the-art (SOTA) solutions from the field of Automatic Speech Recognition (ASR), the sequencing decoding algorithm utilizes a deep neural network trained on vast amounts of data to identify the optimal sequence from all possible combinations and decodes it with precision. The CycloneSEQ has independently developed a basecalling algorithm model. This algorithm model, based on extensive data and large-scale distributed training, achieves a high accuracy rate of 97% and is capable of real-time sequencing decoding for multiple flow cells, demonstrating robust performance.


 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL











 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club




