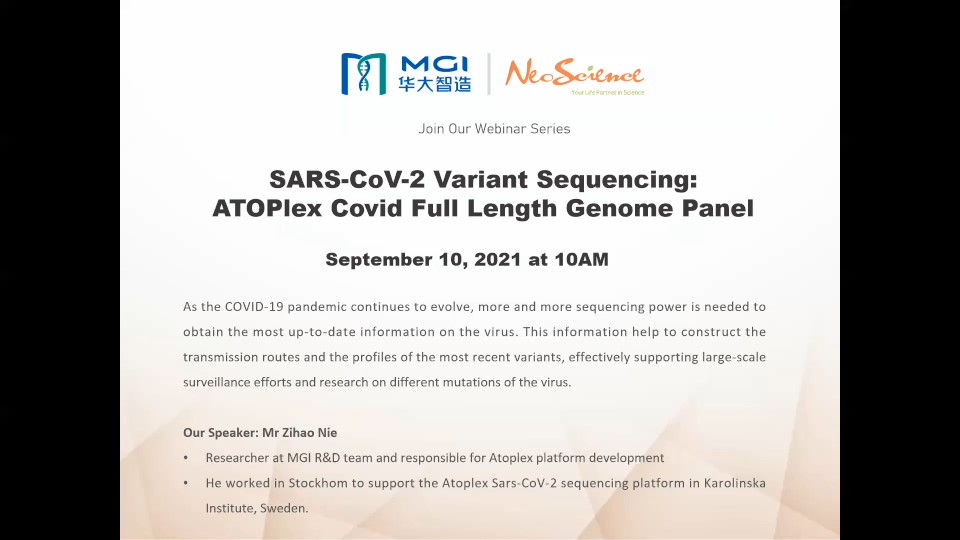
As the COVID-19 pandemic continues to evolve, more and more sequencing power is needed to obtain the most up-to-date information on the virus. These information help to construct the transmission routes and the profiles of the most recent variants, effectively supporting large-scale surveillance efforts and research on different mutations of the virus.
Due to its high-throughput, massively parallel sequencing (MPS) has exhibited enormous capacity and will continue to contribute the surveillance, variation and evolution analysis of SARS-CoV-2.
ATOPlex is a MPS platform that applies MGI proprietary ultra-high multiplex PCR-based enrichment technique in customized panel design. It provides a streamlined workflow for complete genome sequencing and epidemiological study of SARS-CoV-2. After amplifying relevant genomic segments, subsequent sequencing on MGI's DNBSEQ platform achieves highly accurate coronavirus genotyping. The platform also includes ATOPlex RNA Library Prep Set, a reagent that converts viral RNA to sequencing libraries in 5 hours. It is suitable for population-scale virus detection, surveillance and tracing.
If you have any questions, you can send an email to:
MGI_service@mgi-tech.com


 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL











 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club