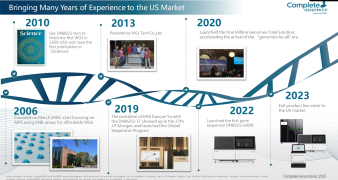
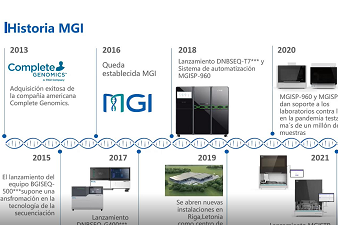
【The Future of Omics Research Summit】Dr. Ashraf Dallol, PhD in Biomedical Science from the University of Ulster and Chief Scientific Director at NoorDx,
Past list
Multiplexed Single Cell Genomics with MGI CoolMPS™ Sequencing Chemistry
Release date:2019-12-17Writer:MGIViews:6184Share
Multiplexed single cell genomics has been used to increase the throughput of droplet based single cell assays enabling cost effective library preparation of hundreds of thousands to millions of single cells. Multiplexing can be performed leveraging natural genetic variants from genetically distinct individuals or cell hashing based on antibody staining or lipid labels to assign single cells to their donor. The success of these methods hinges on the quality of the next generation sequencing data. To assess the feasibility of performing multiplexed single cell genomics with coolMPS sequencing we prepared primary peripheral mononuclear cells (PBMCs) from 3 healthy individuals, and performed CITE-seq (Abseq, BD), cell hashing (TotalSeq-A, BioLegend), and RNA-sequencing using 10X Genomics’ 3’v3 kit solution. The library was sequenced with both an Illumina Novaseq 6000 system and an early alpha version of the CoolMPS™ sequencing kit in development at MGI. Here, we present comparative data on sequencing quality metrics, cell calling, clustering, and cell classification.

Ms. Gracie Gordon
PhD candidate, Biological and Medical Informatics Graduate Program, Department of Medicine and Institute for Human Genetics, UCSFIf you have any questions, you can send an email to:
MGI_marketing@mgi-tech.com


 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL











 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club