MGI has made upgrades in its genetic sequencer MGISEQ-2000 to make it more productive and effective.

What’s New in MGISEQ-2000:
- Average effective reads/flow cell has been increased up to 1800M, meaning increase in data throughput while retaining the same price
- The cost per Gb of data output has been reduced under the same read length
o Sequencing time of read length PE100 has been reduced to only 2 days
o Sequencing throughput has been increased throughout the year
o Now compatible with the dual-index system
- Faster and more flexible
Details on Updated Technologies:
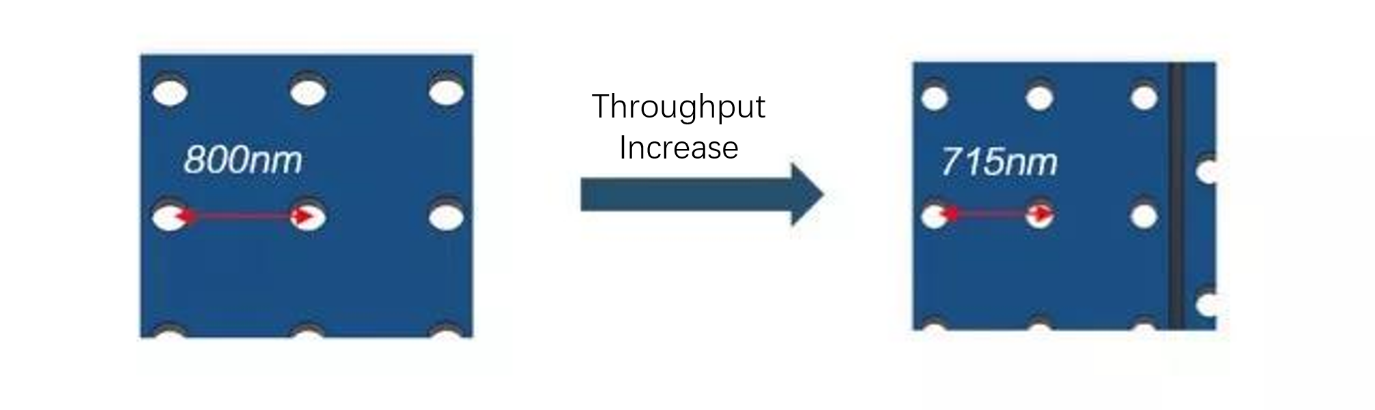
1. Sequencing throughput has been increased through densified flow cell, with the change in pitch from 800nm to 715nm
2. Noise has been reduced in signal between DNBs with the optimization of base-calling algorithm
3. Sequencing time has been shortened by upgrading biochemical reagent
Coming soon: Flow cells and reagents that meet lower throughput requirements
Please stay tuned!
Data talks:
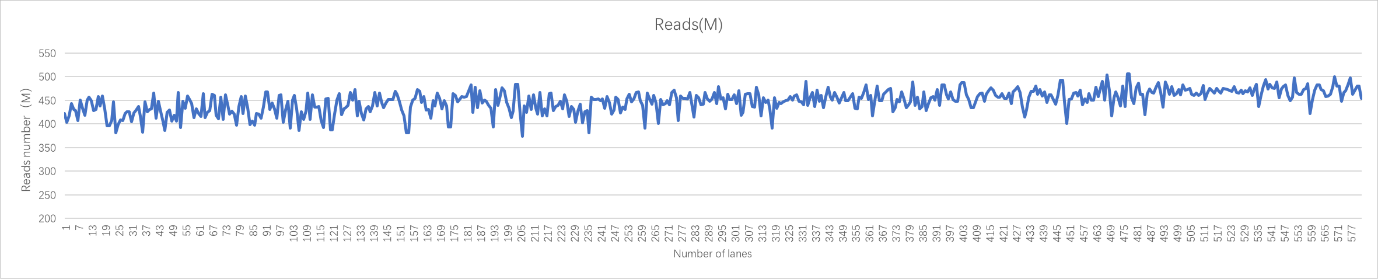
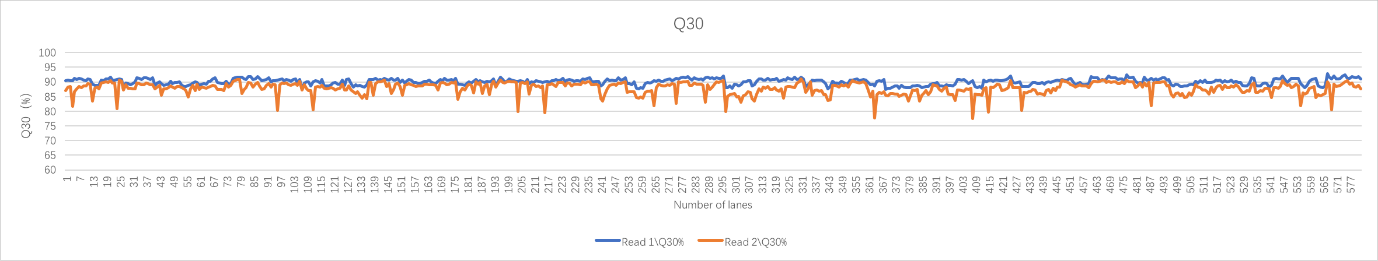
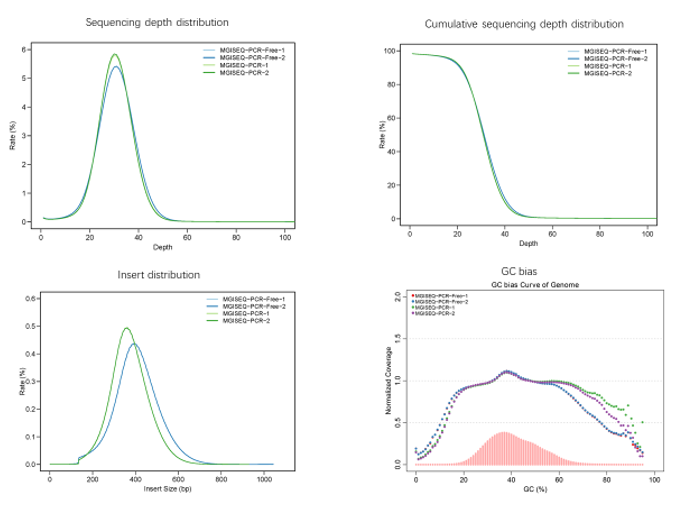
Real sample data generated from 19 upgraded MGISEQ-2000RS gene sequencers, of PE100+10 dual flow cell, whole genome sequencing and transcriptome sequencing of 582 lanes show that data throughput of above 95% lanes are above 400M reads. Q30 of Read 1 is above 87% while the Q30 of Read 2 is above 80% (for 99% lanes).
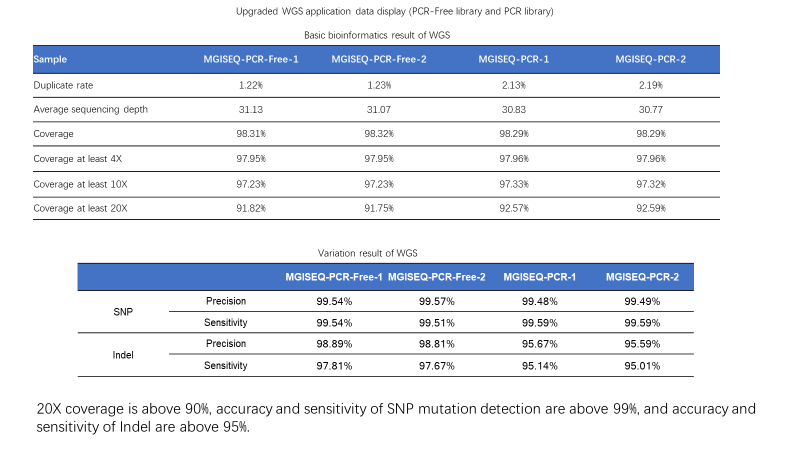
20X coverage is above 90%, accuracy and sensitivity of SNP mutation detection is above 99%, and accuracy and sensitivity of Indel is above 95%.
Sample: NA12878 Bioinformatics pipeline: MegaBOLT Reference: Hg19


 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL











 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club