
Dr. Radoje Drmanac, MGI chief scientific officer, gave a keynote speech on the topic "DNA Nanoballs and Single-Tube LFR: Affordable "Perfect" Genome Sequencing at the AGTA 2018 Conference in Stamford Grand Adelaide, Australia on November 5.

Single tube long fragment read (stLFR) technology enables efficient WGS, haplotyping, and contig scaffolding. It is based on adding the same barcode sequence to sub-fragments of the original DNA molecule (DNA co-barcoding). It uses the surface of microbeads to create millions of miniaturized compartments in a single tube. Using a combinatorial process over 1.8 billion unique barcode sequences were generated on beads, enabling practically non-redundant co-barcoding in reactions with 50 million barcodes.
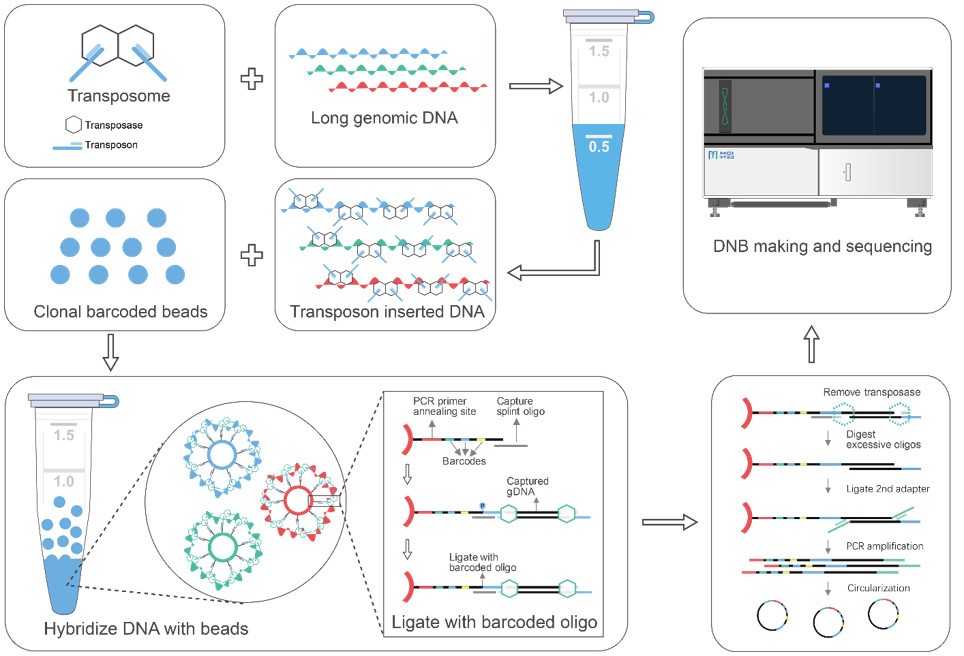
MGI’s newest launched MGIEasy stLFR Library Prep Kit adopts the stLFR technology, it significantly reduces the cost and complexity of long fragment library preparation.
Learn more about MGIEasy stLFR Library Prep Kit here:
https://en.mgitech.cn/Products/reagents_info/id/18
Apply for a trial: http://da9k7j9d8fb7j69h.mikecrm.com/LtFySQU

Read full analysis of stLFR data performed by Ou Wang et al.: https://www.biorxiv.org/content/biorxiv/early/2018/05/17/324392.1.full.pdf


 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL











 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













