Commentary by Dr. Radoje Drmanac, CSO of MGI / Co-Founder of Complete Genomics
This year marks a decade since DNBSEQ technology, painstakingly created and developed by my team and I, was commercialized. As a father and a grandfather myself, DNBSEQ is my other baby – or not so much a baby now. At 10 years old, I imagine it as on the cusp of adolescence, running around with friends and thinking and sounding almost like a “grown-up”.
The last 10 years went by in a blink, and as I reflect on my journey in genomics (which spans far beyond the 10 years), I am filled with pride and awe at how far we as an industry have come.
Making my mark
When I started collage in Serbia studding molecular biology in 1977, the Sanger and Maxam-Gilbert sequencing methods were invented. Then, in 1986, the world welcomed the first commercial DNA sequencer, the ABI Prism 370A, touting a throughput of 1,000 bp per day.
A year later, I invented sequencing by hybridization and soon after DNA microarrays as first massively parallels sequencing (MPS) solution for efficient genome sequencing. In 1991 I moved with my team from Serbia to Argonne National Laboratories as part of Human Genome Project (HGP), which became a pivotal moment that set the stage for the revolutionary advancements we see today. The project was not just about mapping the human genome; it was about unlocking the secrets of life itself. I proposed and developed DNA sequencing-by-hybridization, which laid the groundwork for more efficient and higher-throughput sequencing methods.

Arriving at a turning point
The HGP marked the beginning of a new era in biology, akin to sending humans to the moon. It showed us that large-scale projects were feasible and sparked a demand for sequencing more genomes. The project made shorter reads more usable by providing a reference genome, enabling exome and panel sequencing. It was personally also a “light bulb moment”, inspiring me to start Complete Genomics, with the goal of maximizing the potential of MPS.
In 2005, amidst the arrival of next-generation sequencing, my team and I invented patterned arrays of DNA nanoballs (DNB), which significantly expanded MPS’ capabilities and became DNBSEQ sequencing platform. This technology offers unparalleled accuracy, eliminating clonal errors and index hopping while generating higher signal density. It was the key to Complete Genomics achieving the milestone of sequencing a human genome for $5,000 in 2010, demonstrating that routine, affordable, and accurate whole-genome sequencing was not only possible but also beneficial. Our merger with MGI allowed us to further develop DNBSEQ and introduce the BGISEQ-500, the first commercial sequencer based on DNBSEQ, in 2015.
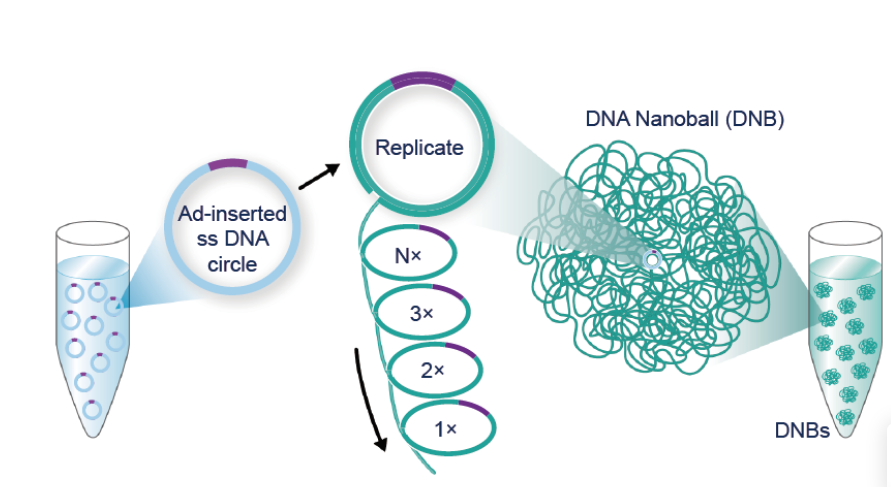
Witnessing the fruits of our endeavors
I recall the watershed moment when we decided to expand beyond just providing whole human genome sequencing services and develop our own commercial sequencer. At the time, we did not have a dedicated engineering or technology development arm. I advised starting an engineering team in MGI, and this led to the hiring of senior engineer Yongwei Zhang, who spoke both English and Chinese to help bridge the gap between the two sites. Yongwei, who has remained in MGI, was instrumental in building the BGISEQ-500. I vividly recall the wave of emotions that swept over me as I saw the machine for the first time, and I was even more astonished to see not just one – but 50 and even 100 of them – in production.
Over the past decade, we have continually pushed the envelope in what DNBSEQ can do to global critical acclaim. To date, users from 100 countries and regions across six continents have published over 10,000+ high-level articles on the DNBSEQ platform, involving more than 5,406 species studied, and having fully verified its advantages in data quality and many other aspects. DNBSEQ now serves as the backbone of MGI's suite of cutting-edge sequencing instruments, including the versatile DNBSEQ-G400 benchtop sequencer, and the ultra-fast DNBSEQ-T7 with a throughput of 1-7 Tb per day. More recently, we also launched the DNBSEQ-T1+, one of the fastest T-level benchtop sequencers globally with the capability of completing a sequencing workflow in only 24 hours.

Indeed, genomics has advanced at an incredible pace, with sequencing costs falling. From the $3 billion spent on the first human genome to sub-$100 genomes made available today by way of our DNBSEQ-T20×2 ultra-high throughput sequencer, this journey has surpassed Moore's Law prediction. I have long envisioned a future where molecular health monitoring becomes an integral part of everyday life. These recent developments in DNBSEQ are making routine genome sequencing for all a more tangible reality than ever. My team and I are determined to reach a $10 genome through advancements like increased DNB array density, our single-tube long fragment read (stLFR) technology, CycloneSEQ technology and other novel technologies, enabling affordable fully phased, highly accurate and complete whole-genome sequencing for health monitoring, personalized medicine, and other applications.
Welcoming a future of unlimited possibilities
The journey from when nucleic acids were first isolated in 1869 to the HGP, and the current era of genomics has been a testament to human ingenuity and collaboration. The applications of sequencing technologies are vast and ever-expanding. I look forward to celebrating 10 years of the DNBSEQ-G400 or 10 years of the DNBSEQ-T7, and I’m excited about what these platforms will bring to genomics and life science in the coming years.

In this past decade since DNBSEQ was commercialized, it has not only transformed the field of genomics but has also empowered numerous national genome projects around the world and paved the way for improved global health. As DNBSEQ continues to evolve, we prepare to leverage its potential and ring in a new era where genomics will touch every aspect of our lives, promising a future where health and longevity are within our grasp. This year, we are not just celebrating a milestone; we are embracing the limitless possibilities of DNBSEQ that lie ahead.


 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL











 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













