Last month, Wang Jian, chairman of MGI, went to the frontlines to support prevention and control of the 2019-nCoV outbreak in Wuhan. "Precise nucleic acid testing is an important basis for clinical diagnosis and discharge of recovered patients, as well as ending isolation of contacts,” he said, pointing to the urgent need for a testing center that can process a large volume of samples daily.
On February 5, the Huo-Yan Laboratory, an emergency test laboratory run by MGI and designed to handle 10,000 samples daily to detect 2019-nCoV, officially started trial operation in Wuhan. The lab will greatly accelerate the testing of novel coronavirus cases so that suspected infections can be eliminated or confirmed as soon as possible. MGI's high-throughput automated equipment and sequencers, tested in epidemic detection of novel coronavirus, will also be put into use in the Huo-Yan Laboratory.

Wuhan Huo-Yan Laboratory officially put into trial operation
MGI's novel coronavirus detection package:Automated nucleic acid extraction + high-throughput sequencing
With epidemic prevention and control at a critical stage, accurate detection is critical for the diagnosis of patients with fever, screening of high-risk populations and screening of suspected cases for "early detection, isolation, diagnosis, and treatment".
The real-time quantitative reverse polymerase chain reaction (RT-PCR) method of novel coronavirus detection currently applied in the frontlines can screen the virus and is suitable for rapid diagnosis of the presence of coronavirus in patient samples. High-throughput genetic sequencing technology is more comprehensive. Besides being able to diagnose the presence of coronavirus, the evolutionary source of the virus, the pathogenic and pathological mechanism and the mutation of the virus can also be studied and analyzed. High-throughput sequencing can also detect all pathogens, including the novel coronavirus and the entire gene sequence of the pathogen. It is of great significance for the diagnosis and treatment of critically ill patients. In order to achieve efficient and accurate detection, it is urgent to build capacity for nucleic acid detection and supporting sequencing capabilities.
In response to the needs of front-line epidemic detection, MGI provides the 2019-nCoV detection package, which not only provides a one-stop automated nucleic acid extraction package for real-time quantitative reverse polymerase chain reaction (RT-PCR), but also provides a flexible sequencer for different throughput needs.
In addition to its application in the Huo-Yan Laboratory in Wuhan, this package has also been implemented to varying degrees in China's National Centers for Disease Control (CDC) and various Disease Control Departments to speed up the detection and diagnosis of suspected cases.
Viral nucleic acid automated extraction: One-stop sample processing laboratory to accelerate virus detection
At present, in the detection of the novel coronavirus, virus nucleic acid extraction of suspected samples is mostly performed manually, which has become the biggest problem limiting the speed of sample detection and increasing the risk of virus infection in frontline medical staff.

In order automate this process, MGI offers two automated sample preparation systems, MGISP-100 and MGISP-960, that can be used with virus RNA extraction reagents and consumables. It has flexible throughput and fast automation, and is highly expandable. No manual intervention is needed after the machine is turned on. The system can complete automated nucleic acid extraction in one hour, improving safety and control.
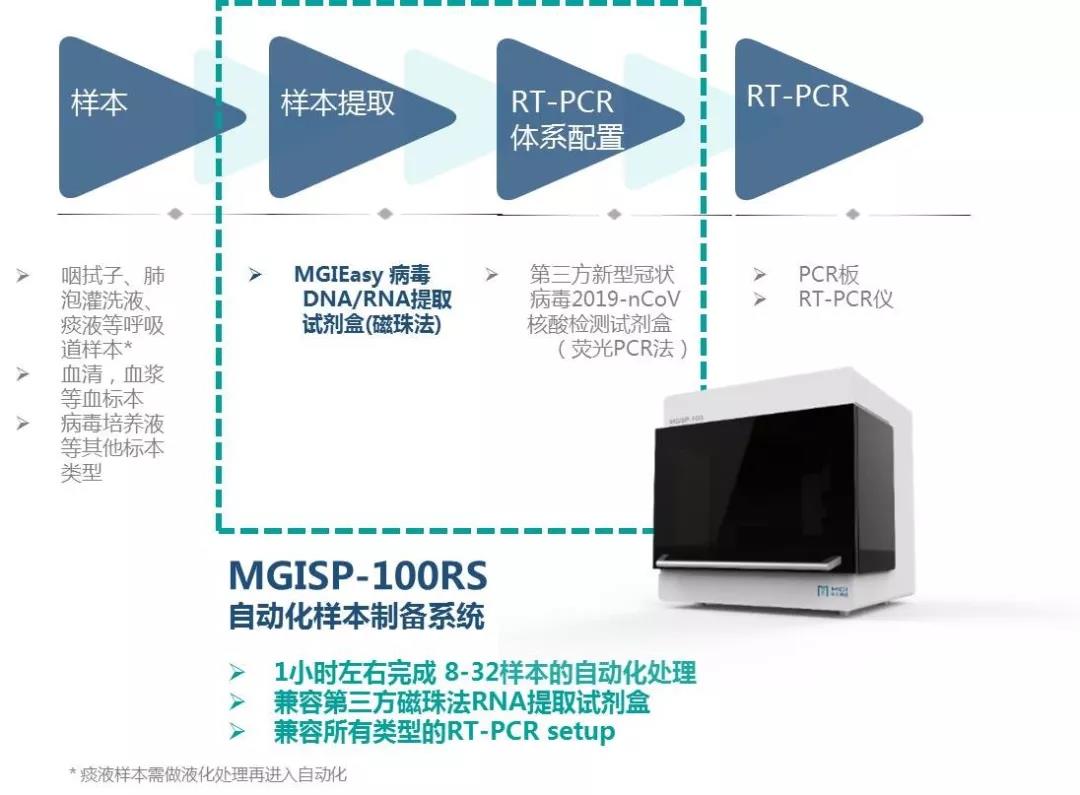
MGISP-100 is the world's first automated sample preparation system to receive a Class II medical device registration certificate. It can extract 8-32 samples in 40-80 minutes, which can reduce the manual operation time of medical personnel by about 2 hours per run and is suitable for small and medium-sized laboratories.
MGISP-960 is a high-throughput automated sample preparation system that has obtained NMPA certification. With a higher degree of automation, it can complete 36-96 samples per hour, and one person can operate two units at the same time, improving efficiency. MGISP-960 is suitable for enterprises, high-throughput users in clinical test settings, hospitals and others. To meet the urgent needs of large-scale populations, the throughput of MGISP-960 can be upgraded to complete 192 samples within 80 minutes.
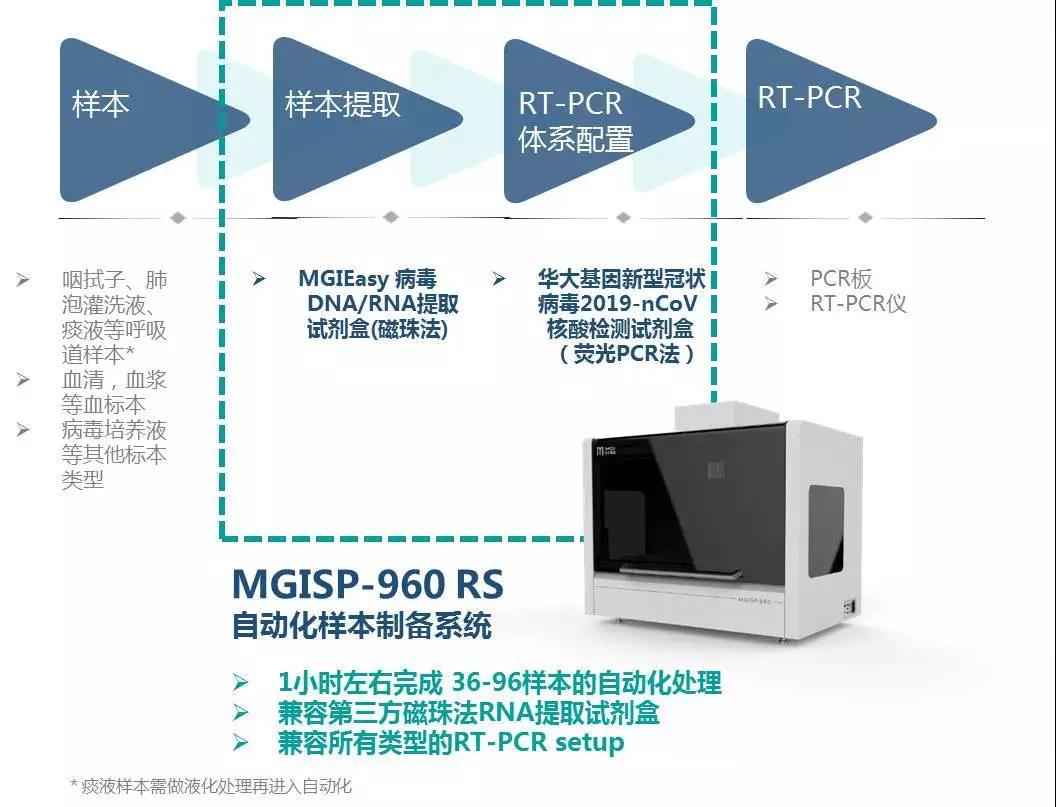
In addition, the program is also equipped with ultraviolet and HEPA air filtration systems, which can be used for daily sterilization and cleaning, and can be adapted to the current mainstream novel coronavirus nucleic acid detection kit (fluorescent PCR method), and third-party magnetic bead extraction reagents.
High-throughput sequencing: Improving accuracy and monitoring mutations

At present, the traceability, monitoring of mutations, molecular epidemiology, and pathogenesis of human infections of the novel coronavirus needs to be further clarified. The virus may mutate during transmission and will affect the sensitivity of RT-PCR, which can be compensated by using high-throughput sequencing, thus possible virus mutations can be detected. Some samples with lower virus content use high-throughput sequencing when the content is lower than the minimum detection limit of RT-PCR. The sequencing method can improve the successful detection rate and avoid missed detection and false detection. For suspected infections, multiple infections or secondary infections, high-throughput sequencing can be used for concurrent detection, providing more information about the pathogens of possible infections.
High-throughput sequencing methods can be used to effectively deal with problems from rapid detection of unknown pathogens, to rapid identification of known sequences, to confirmation of positive sample results, and dynamic monitoring of possible sequence variations. MGI 2019-nCoV detection package includes a number of sequencers that have been approved by the National Medical Products Administration and can be used to clinically diagnose other coronaviruses and respiratory tract infections, including novel coronaviruses. It can also be used to quickly detect viral sequences, and to monitor possible virus mutations and provide tools and platform support.
MGI's high-throughput sequencing packages can meet the needs of various sample sizes, sequencing data volumes and throughput requirements, from the DNBSEQ-G series (DNBSEQ-G50, DNBSEQ-G400/DNBSEQ-G400 FAST), to DNBSEQ-T series (DNBSEQ-T7) and other models for epidemic prevention and control.
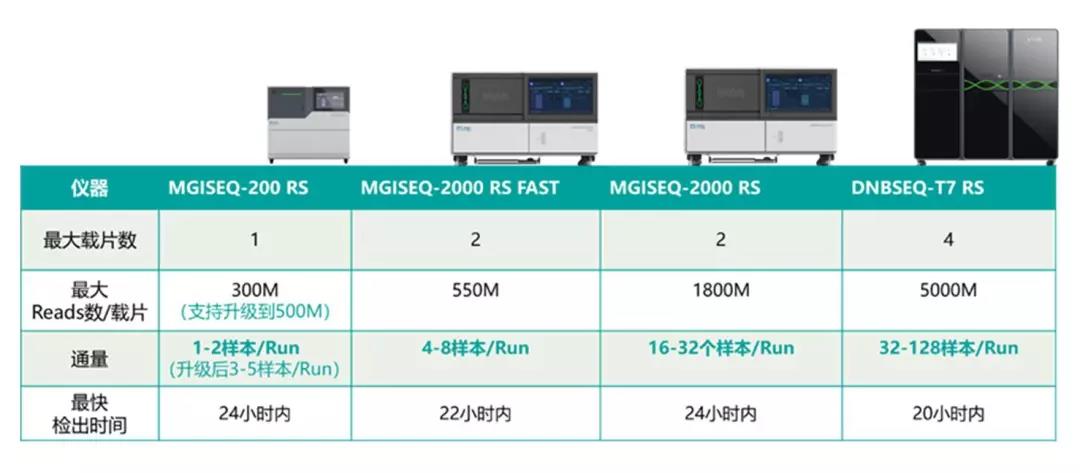
For analyzing individual samples with unknown fever, unknown etiology, and diagnosable disease, the DNBSEQ-G series sequencer (DNBSEQ-G50, DNBSEQ-G400/DNBSEQ-G400 FAST and other models) can quickly identify them within 24 hours, providing data support for sequence analysis and virus evolution.
For analyzing multiple suspected samples, the DNBSEQ-G400 platform is effective. Its single operation supports 32 samples on the machine at the same time. When facing a large number of suspected samples, DNBSEQ-T7 is an effective tool for rapid identification. The highest throughput sequencer to date, DNBSEQ-T7 can support 128 samples for sequencing within 5-6 hours (sequencing strategy: SE50). It can go from sample to data analysis results in 20 hours or less.
DNBSEQ-G400 and DNBSEQ-G50 remain their original product names as MGISEQ-2000 and MGISEQ-200 respectively in China and some other overseas countries.


 Sequencer Products: SEQ ALL
Sequencer Products: SEQ ALL











 Technologies
Technologies Applications
Applications Online Resources
Online Resources Data Bulletins
Data Bulletins Service & Support
Service & Support Global Programs
Global Programs Introduction
Introduction Newsroom
Newsroom Doing Business With Us
Doing Business With Us Creative Club
Creative Club













