DCS Lab
Empower Global Cutting-edge Genomics Research



Essential DCS Research Exhibition
*Data updated to December, 2023
Establishing the Foundation of State-of-art Research
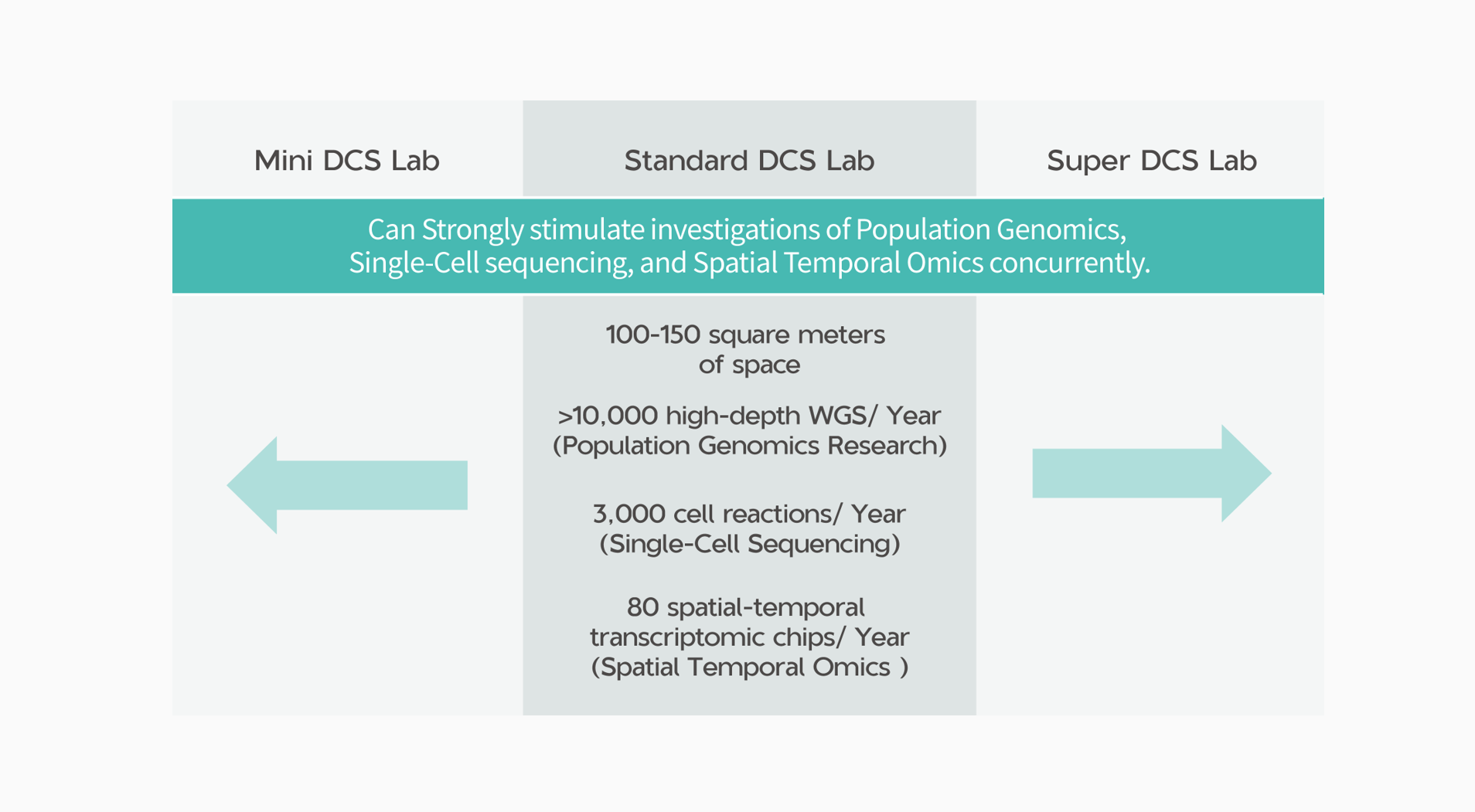
*The number of instrument units and the usage of reagents and consumables based on the specific size and scale of your experiment.
Videos | T20×2: An Ultra-High Throughput SequencerNews | National genomic sequencing project in BrazilNews | MGI Empowers the Completion of Nearly 60,000 Samples for The Million Microbiome of Humans Project
Publication List | Single Cell Sequencing (2020-2023)- incomplete statisticsVideos | DNBelab C4 Pocket Single-Cell Lab
News | International Scientists Release Most Complete Primate Brain Cell Atlas to DateNews | Scientists build world's first spatio-temporal atlas of brain regeneration by STOmics Stereo-seq